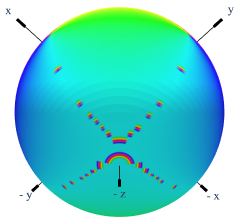
I’m plotting a cyclic value over a sphere using graph_objects.Surface, and I’m using the ‘Phase’ colourmap. When the value goes from 1->0, the plot interpolates through the whole colormap between the two values as shown below.

I was wondering if there was a way to disable the interpolation, or interpolate cyclic colormaps correctly.
I have also tried plotting as Mesh3d and using intensitymode=‘cell’, which looks roughly correct, but then I run into a new set of problems as I have n-long 1D lists of x, y, z, and phase, and I’m getting confused about how to convert the n vertex values to ~2n face values.
Any help is appreciated, thank you!