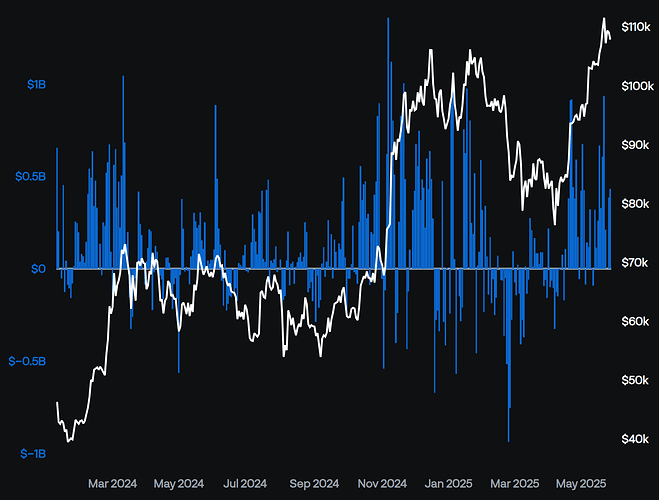
I have a Pandas DataFrame that has a DatetimeIndex with a business day frequency. When I plot it using rangebreaks the x-axis labels look fine when the entire range is shown, but they appear sparse when zoomed in to a few weeks or less. I don’t know the logic behind how these labels are selected, but I’m wondering if there’s a better way to show these labels programmatically? Ideally beyond a certain selected range threshold (~20 points), it would switch to showing labels at every point or every other point. I don’t mind which, as long as there wasn’t a cluster of labels as can be seen in the 2nd screenshot below.
I would like to keep the axis as a datetime type so that Plotly can still auto-adjust the tick density (the full range is a few hundred data points).
Here is a .tail(20) of my data, showing the DatetimeIndex frequency:
Date
2025-05-01 422500000.0
2025-05-02 674900000.0
2025-05-05 425500000.0
2025-05-06 -85700000.0
2025-05-07 142300000.0
2025-05-08 117400000.0
2025-05-09 321400000.0
2025-05-12 5200000.0
2025-05-13 -91400000.0
2025-05-14 319500000.0
2025-05-15 114900000.0
2025-05-16 260200000.0
2025-05-19 667400000.0
2025-05-20 329200000.0
2025-05-21 607100000.0
2025-05-22 934800000.0
2025-05-23 211700000.0
2025-05-26 0.0
2025-05-27 385400000.0
2025-05-28 432700000.0
Freq: B, Name: Total, dtype: float64
This is a plot of the full range:
and this is a zoomed-in view. Here is where the x-axis labels become sparse, and I can’t understand the logic behind choosing these labels:
If it matters, I am defining a figure via make_subplots.